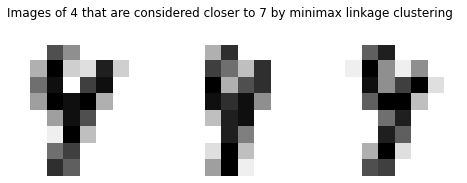Examples¶
It is recommended that you go through the quick start guide before reading this page.
This page is generated by a Jupyter notebook which can be opened and run in Binder or Google Colab by clicking on the above badges. To run it in Google Colab, first you need to install PyMinimax and a newer version of scikit-learn in Colab:
[ ]:
!pip install pyminimax scikit-learn==0.23
Random Points in 2D¶
In this example we perform minimax linkage clustering on a toy dataset of 20 random points in 2D:
[1]:
import numpy as np
from pandas import DataFrame
np.random.seed(0)
X = np.random.rand(20, 2)
DataFrame(X, columns=['x', 'y'])
[1]:
| x | y | |
|---|---|---|
| 0 | 0.548814 | 0.715189 |
| 1 | 0.602763 | 0.544883 |
| 2 | 0.423655 | 0.645894 |
| 3 | 0.437587 | 0.891773 |
| 4 | 0.963663 | 0.383442 |
| 5 | 0.791725 | 0.528895 |
| 6 | 0.568045 | 0.925597 |
| 7 | 0.071036 | 0.087129 |
| 8 | 0.020218 | 0.832620 |
| 9 | 0.778157 | 0.870012 |
| 10 | 0.978618 | 0.799159 |
| 11 | 0.461479 | 0.780529 |
| 12 | 0.118274 | 0.639921 |
| 13 | 0.143353 | 0.944669 |
| 14 | 0.521848 | 0.414662 |
| 15 | 0.264556 | 0.774234 |
| 16 | 0.456150 | 0.568434 |
| 17 | 0.018790 | 0.617635 |
| 18 | 0.612096 | 0.616934 |
| 19 | 0.943748 | 0.681820 |
Below is the dendrogram.
[2]:
import matplotlib.pyplot as plt
from pyminimax import minimax
from scipy.spatial.distance import pdist
from scipy.cluster.hierarchy import dendrogram
Z = minimax(pdist(X), return_prototype=True)
plt.figure(figsize=(10, 4))
dendrogram(Z[:, :4])
plt.show()
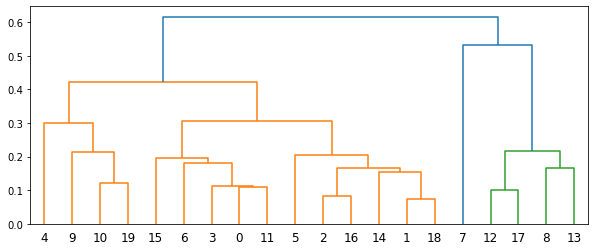
A unique advantage of minimax linkage hierarchical clustering is that every cluster has a prototype selected from the original data. This is a representative data point of the cluster.
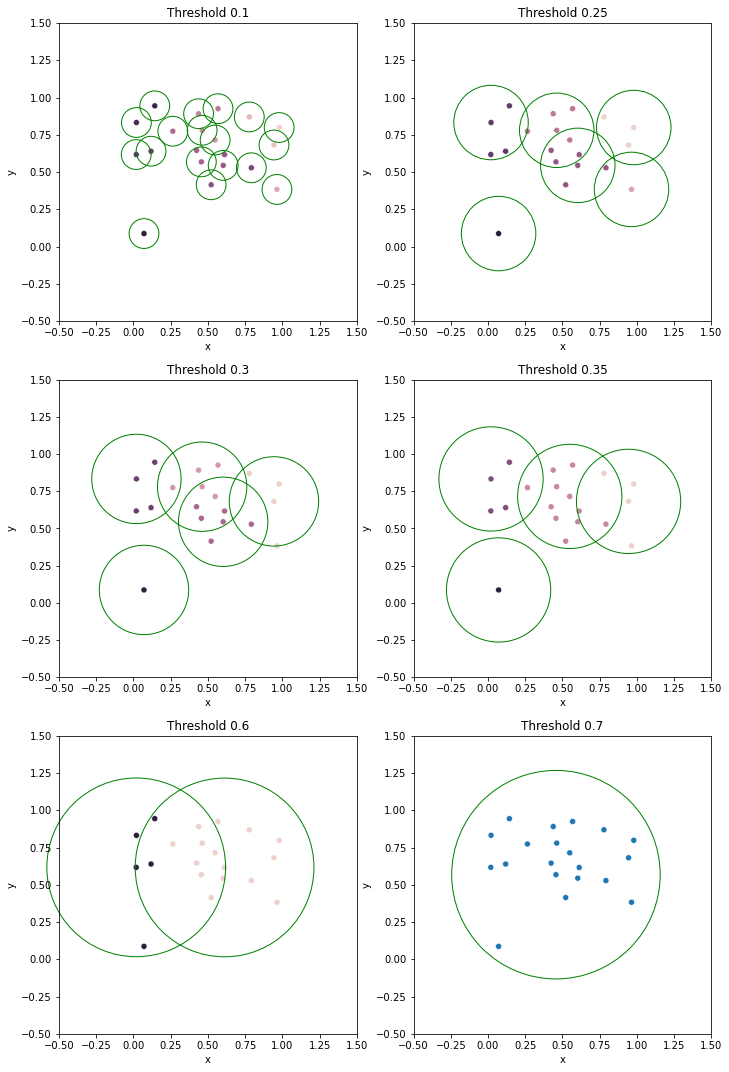
The threshold used to cut the dendrogram is also interpretable. Suppose the dendrogram is cut at threshold \(t\), splitting the data into clusters \(G_1, G_2, \ldots\) with corresponding prototypes \(p_1, p_2, \ldots\). Then, for any \(i\), all data points in \(G_i\) must be in the circle centered at \(p_i\) with radius \(t\). That is, the distance from the prototype of a cluster to any data point in the same cluster must be less than or equal to \(t\).
Here we draw the clusters and the circles for various thresholds. The data points at the center of the circles are the prototypes.
[3]:
import seaborn as sns
from pandas import DataFrame
from pyminimax import fcluster_prototype
cuts = [0.1, 0.25, 0.3, 0.35, 0.6, 0.7]
fig, axs = plt.subplots(3, 2, figsize=(10, 15))
for ax, cut in zip(axs.ravel(), cuts):
clust_proto = fcluster_prototype(Z, t=cut, criterion='distance')
df = DataFrame(np.concatenate([X, clust_proto], axis=1), columns=['x', 'y', 'clust', 'proto'])
sns.scatterplot(data=df, x='x', y='y', hue='clust', legend=None, ax=ax)
ax.set(xlim=(-0.5, 1.5), ylim=(-0.5, 1.5), aspect=1, title=f'Threshold {cut}')
protos = np.unique(df['proto'].map(int).values)
for proto in protos:
circle = plt.Circle(X[proto], cut, edgecolor='g', facecolor='none', clip_on=False)
ax.add_patch(circle)
fig.tight_layout()
plt.show()
Hand-Written Digits¶
In this example we perform minimax linkage clustering on images of hand-written digits 1, 4 and 7. The data we use is a subset of the scikit-learn hand-written digit images data. The below code from its documentation prints the first few images in this dataset.
[4]:
import matplotlib.pyplot as plt
from sklearn import datasets
digits = datasets.load_digits()
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, label in zip(axes, digits.images, digits.target):
ax.set_axis_off()
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
ax.set_title('Training: %i' % label)
First we load the data in a pandas DataFrame, and filter out images that are not 1, 4 or 7. The resulting DataFrame digits147 has 542 rows, each having 65 values. The first 64 are a flattened \(8\times 8\) matrix representing the image, and the last value in the target column indicates this image is a 1, 4 or 7.
[5]:
digits = datasets.load_digits(as_frame=True)['frame']
digits147 = digits[digits['target'].isin([1, 4, 7])].reset_index(drop=True)
digits147
[5]:
| pixel_0_0 | pixel_0_1 | pixel_0_2 | pixel_0_3 | pixel_0_4 | pixel_0_5 | pixel_0_6 | pixel_0_7 | pixel_1_0 | pixel_1_1 | ... | pixel_6_7 | pixel_7_0 | pixel_7_1 | pixel_7_2 | pixel_7_3 | pixel_7_4 | pixel_7_5 | pixel_7_6 | pixel_7_7 | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 | 12.0 | 13.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 11.0 | 16.0 | 10.0 | 0.0 | 0.0 | 1 |
| 1 | 0.0 | 0.0 | 0.0 | 1.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 16.0 | 4.0 | 0.0 | 0.0 | 4 |
| 2 | 0.0 | 0.0 | 7.0 | 8.0 | 13.0 | 16.0 | 15.0 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 13.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7 |
| 3 | 0.0 | 0.0 | 0.0 | 0.0 | 14.0 | 13.0 | 1.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 13.0 | 16.0 | 1.0 | 0.0 | 1 |
| 4 | 0.0 | 0.0 | 0.0 | 8.0 | 15.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 15.0 | 4.0 | 0.0 | 0.0 | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 537 | 0.0 | 0.0 | 0.0 | 1.0 | 13.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 15.0 | 7.0 | 0.0 | 0.0 | 4 |
| 538 | 0.0 | 0.0 | 3.0 | 10.0 | 16.0 | 16.0 | 4.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 3.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7 |
| 539 | 0.0 | 1.0 | 10.0 | 16.0 | 15.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 10.0 | 15.0 | 2.0 | 0.0 | 0.0 | 0.0 | 7 |
| 540 | 0.0 | 0.0 | 0.0 | 1.0 | 12.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 14.0 | 9.0 | 0.0 | 0.0 | 4 |
| 541 | 0.0 | 0.0 | 0.0 | 3.0 | 15.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 16.0 | 4.0 | 0.0 | 0.0 | 4 |
542 rows × 65 columns
For example, the first 64 values of the first row is the below matrix flattened. This is a matrix of grayscale values representing an image of 1.
[6]:
digits147.iloc[0].values[:-1].reshape(8, 8)
[6]:
array([[ 0., 0., 0., 12., 13., 5., 0., 0.],
[ 0., 0., 0., 11., 16., 9., 0., 0.],
[ 0., 0., 3., 15., 16., 6., 0., 0.],
[ 0., 7., 15., 16., 16., 2., 0., 0.],
[ 0., 0., 1., 16., 16., 3., 0., 0.],
[ 0., 0., 1., 16., 16., 6., 0., 0.],
[ 0., 0., 1., 16., 16., 6., 0., 0.],
[ 0., 0., 0., 11., 16., 10., 0., 0.]])
We drop the target column from the data, compute the extended linkage matrix and draw the dendrogram.
[7]:
from pyminimax import minimax
from scipy.spatial.distance import pdist
from scipy.cluster.hierarchy import dendrogram
X = digits147.drop('target', axis=1).values
Z = minimax(pdist(X), return_prototype=True)
plt.figure(figsize=(10, 4))
dendrogram(Z[:, :4])
plt.show()
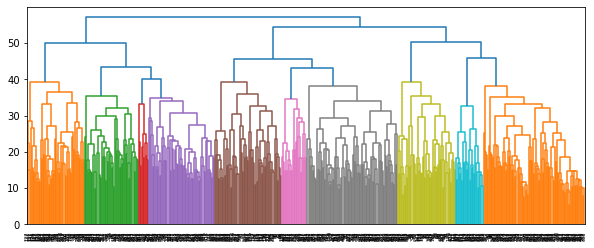
The 3rd column of the extended linkage matrix is the distance between the two clusters to be merged in each row. The 3rd last merge has distance 50.3388, indicating that if the dendrogram is cut at a threshold slightly above 50.3388, there will be 3 clusters.
The format of the extended linkage matrix is detailed in the quick start guide and the Scipy documentation.
[8]:
from pandas import DataFrame
DataFrame(Z[-3:, :], columns=['x', 'y', 'distance', 'n_pts', 'prototype'])
[8]:
| x | y | distance | n_pts | prototype | |
|---|---|---|---|---|---|
| 0 | 1072.0 | 1078.0 | 50.338852 | 182.0 | 135.0 |
| 1 | 1077.0 | 1080.0 | 54.488531 | 360.0 | 137.0 |
| 2 | 1079.0 | 1081.0 | 57.122675 | 542.0 | 22.0 |
The cluster and prototypes is computed by pyminimax.fcluster_prototype and put together with the target column. The result is sorted by target for better visualization. As expected, most of the images of 1 are in the same cluster (cluster #3), and most of the images of 7 are in a different cluster (cluster #1).
[9]:
from pyminimax import fcluster_prototype
clust, proto = fcluster_prototype(Z, t=52, criterion='distance').T
res = digits147.assign(clust=clust, proto=proto)
res = res[['target', 'clust', 'proto']].sort_values(by='target')
res
[9]:
| target | clust | proto | |
|---|---|---|---|
| 0 | 1 | 3 | 135 |
| 154 | 1 | 3 | 135 |
| 157 | 1 | 3 | 135 |
| 160 | 1 | 3 | 135 |
| 379 | 1 | 3 | 135 |
| ... | ... | ... | ... |
| 414 | 7 | 1 | 341 |
| 126 | 7 | 1 | 341 |
| 314 | 7 | 1 | 341 |
| 138 | 7 | 1 | 341 |
| 136 | 7 | 1 | 341 |
542 rows × 3 columns
An even better visualization is the below heat map of the target column and the cluster column. It is clear that all images of 1 are in cluster #3, all images of 7 are in cluster #1, and most of images of 4 are in cluster #2. There are only a few images of 4 wrongly put into cluster #1. That is, minimax linkage clustering finds those images closer to 7.
[10]:
import seaborn as sns
sns.heatmap(res[['target', 'clust']]);
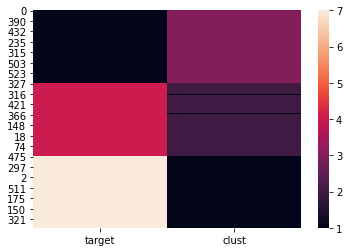
The prototypes are the 135th, 341st, and 464th row of the original DataFrame digits147.
[11]:
import numpy as np
protos = np.unique(res['proto'])
protos
[11]:
array([135, 341, 464], dtype=int32)
We print out the images of the prototypes. These are the representative images of 1, 4 and 7.
[12]:
fig, axs = plt.subplots(nrows=1, ncols=3, figsize=(8, 3))
for ax, proto, label in zip(axs, protos[[0, 2, 1]], [1, 4, 7]):
ax.set_axis_off()
image = digits147.drop('target', axis=1).iloc[proto].values.reshape(8, 8)
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')
ax.set_title(f'Prototype of {label}s')
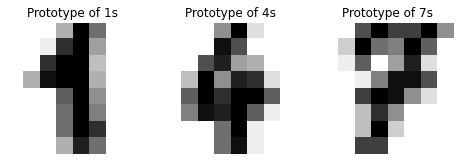
There are 3 images of 4 considered closer to 7. Their indices are 482, 488 and 501, given which we can print out the images for inspection.
[13]:
res[(res['target']==4) & (res['clust']==1)]
[13]:
| target | clust | proto | |
|---|---|---|---|
| 501 | 4 | 1 | 341 |
| 488 | 4 | 1 | 341 |
| 482 | 4 | 1 | 341 |
Arguably they are indeed closer to 7’s prototype than 4’s.
[14]:
fig, axs = plt.subplots(nrows=1, ncols=3, figsize=(8, 3))
plt.suptitle("Images of 4 that are considered closer to 7 by minimax linkage clustering")
for ax, idx in zip(axs, [501, 488, 482]):
ax.set_axis_off()
image = digits147.drop('target', axis=1).iloc[idx].values.reshape(8, 8)
ax.imshow(image, cmap=plt.cm.gray_r, interpolation='nearest')